navis.connectivity_similarity¶
- navis.connectivity_similarity(adjacency, metric='vertex_normalized', threshold=None, n_cores=1, **kwargs)[source]¶
Calculate connectivity similarity.
This functions offers a selection of metrics to compare connectivity:
Metric
Explanation
cosine
Cosine similarity (see here)
rank_index
Normalized difference in rank of synaptic partners.
matching_index
Number of shared partners divided by total number of partners.
matching_index_synapses
Number of shared synapses (i.e. number of connections from/onto the same partners) divided by total number of synapses. Attention: this metric is tricky when there is a disparity of total number of connections between neuron A and B. For example, consider 100/200 and 1/50 shared/total synapse: 101/250 results in a fairly high matching index of 0.404.
matching_index_weighted_synapses
Similar to matching_index_synapses but slightly less prone to above mentioned error as it uses the percentage of shared synapses:
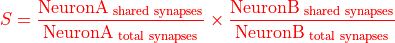
vertex
Matching index that rewards shared and punishes non-shared partners. Based on Jarrell et al., 2012:

Final score is the sum of
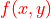 over all edges x, y between
neurons A+B and their partners. C1 determines how negatively a case
where one edge is much stronger than another is punished. C2
determines the point where the similarity switches from negative to
positive. C1 and C2 default to 0.5 and 1, respectively, but can be
changed by passing them in a dictionary as
over all edges x, y between
neurons A+B and their partners. C1 determines how negatively a case
where one edge is much stronger than another is punished. C2
determines the point where the similarity switches from negative to
positive. C1 and C2 default to 0.5 and 1, respectively, but can be
changed by passing them in a dictionary as **kwargs.vertex_normalized
This is vertex similarity normalized by the lowest (hypothetical total dissimilarity) and highest (all edge weights the same) achievable score.
- Parameters:
adjacency (pandas DataFrame | numpy array) – (N, M) observation vector with M observations for N neurons - e.g. an adjacency matrix. Will calculate similarity for all rows using the columns as observations.
metric ('cosine' | 'rank_index'| 'matching_index' | 'matching_index_synapses' | 'matching_index_weighted_synapses' | 'vertex' | 'vertex_normalized') – Metric used to compare connectivity. See notes for detailed explanation.
threshold (int, optional) – Connections weaker than this will be set to zero.
n_cores (int) – Number of parallel processes to use. Defaults to half the available cores.
- Returns:
Pandas DataFrame with similarity scores. Neurons without any connectivity will show up with
np.nanfor scores.- Return type:
DataFrame