navis.plot2d¶
- navis.plot2d(x, method='3d', **kwargs)[source]¶
Generate 2D plots of neurons and neuropils.
The main advantage of this is that you can save plot as vector graphics.
Important
This function uses matplotlib which “fakes” 3D as it has only very limited control over layering objects in 3D. Therefore neurites are not necessarily plotted in the right Z order. This becomes especially troublesome when plotting a complex scene with lots of neurons criss-crossing. See the
methodparameter for details. All methods use orthogonal projection.- Parameters:
x (TreeNeuron | MeshNeuron | NeuronList | Volume | Dotprops | np.ndarray) –
Objects to plot:
multiple objects can be passed as list (see examples)
numpy array of shape (n,3) is intepreted as points for scatter plots
method ('2d' | '3d' (default) | '3d_complex') –
Method used to generate plot. Comes in three flavours:
’2d’ uses normal matplotlib. Neurons are plotted on top of one another in the order their are passed to the function. Use the
viewparameter (below) to set the view (default = xy).’3d’ uses matplotlib’s 3D axis. Here, matplotlib decide the depth order (zorder) of plotting. Can change perspective either interacively or by code (see examples).
’3d_complex’ same as 3d but each neuron segment is added individually. This allows for more complex zorders to be rendered correctly. Slows down rendering though.
soma (bool, default=True) – Plot soma if one exists. Size of the soma is determined by the neuron’s
.soma_radiusproperty which defaults to the “radius” column forTreeNeurons.connectors (bool, default=True) – Plot connectors.
connectors_only (boolean, default=False) – Plot only connectors, not the neuron.
cn_size (int | float, default = 1) – Size of connectors.
linewidth (int | float, default=.5) – Width of neurites. Also accepts alias
lw.linestyle (str, default='-') – Line style of neurites. Also accepts alias
ls.autoscale (bool, default=True) – If True, will scale the axes to fit the data.
scalebar (int | float | str | pint.Quantity, default=False) – Adds scale bar. Provide integer, float or str to set size of scalebar. Int|float are assumed to be in same units as data. You can specify units in as string: e.g. “1 um”. For methods ‘3d’ and ‘3d_complex’, this will create an axis object.
ax (matplotlib ax, default=None) – Pass an ax object if you want to plot on an existing canvas. Must match
method- i.e. 2D or 3D axis.figsize (tuple, default=(8, 8)) – Size of figure.
color (None | str | tuple | list | dict, default=None) – Use single str (e.g.
'red') or(r, g, b)tuple to give all neurons the same color. Uselistof colors to assign colors:['red', (1, 0, 1), ...]. Use ``dictto map colors to neuron IDs:{id: (r, g, b), ...}.palette (str | array | list of arrays, default=None) – Name of a matplotlib or seaborn palette. If
coloris not specified will pick colors from this palette.color_by (str | array | list of arrays, default = None) – Can be the name of a column in the node table of
TreeNeuronsor an array of (numerical or categorical) values for each node. Numerical values will be normalized. You can control the normalization by passing avminand/orvmaxparameter.shade_by (str | array | list of arrays, default=None) – Similar to
color_bybut will affect only the alpha channel of the color. Ifshade_by='strahler'will compute Strahler order if not already part of the node table (TreeNeurons only). Numerical values will be normalized. You can control the normalization by passing asminand/orsmaxparameter.alpha (float [0-1], default=.9) – Alpha value for neurons. Overriden if alpha is provided as fourth value in
color(rgb*a*). You can override alpha value for connectors by usingcn_alpha.clusters (list, default=None) – A list assigning a cluster to each neuron (e.g.
[0, 0, 0, 1, 1]). Overridescolorand usespaletteto generate colors according to clusters.depth_coloring (bool, default=False) – If True, will color encode depth (Z). Overrides
color. Does not work withmethod = '3d_complex'.depth_scale (bool, default=True) – If True and
depth_coloring=Truewill plot a scale.cn_mesh_colors (bool, default=False) – If True, will use the neuron’s color for its connectors.
group_neurons (bool, default=False) – If True, neurons will be grouped. Works with SVG export (not PDF). Does NOT work with
method='3d_complex'.scatter_kws (dict, default={}) – Parameters to be used when plotting points. Accepted keywords are:
sizeandcolor.view (tuple, default = ("x", "y")) – Sets view for
method='2d'.orthogonal (bool, default=True) – Whether to use orthogonal or perspective view for methods ‘3d’ and ‘3d_complex’.
volume_outlines (bool | "both", default=True) – If True will plot volume outline with no fill. Only works with method=”2d”.
dps_scale_vec (float) – Scale vector for dotprops.
rasterize (bool, default=False) – Neurons produce rather complex vector graphics which can lead to large files when saving to SVG, PDF or PS. Use this parameter to rasterize neurons and meshes/volumes (but not axes or labels) to reduce file size.
Examples
>>> import navis >>> import matplotlib.pyplot as plt
Plot list of neurons as simple 2d
>>> nl = navis.example_neurons() >>> fig, ax = navis.plot2d(nl, method='2d', view=('x', '-y')) >>> plt.show()

Add a volume
>>> vol = navis.example_volume('LH') >>> fig, ax = navis.plot2d([nl, vol], method='2d', view=('x', '-y')) >>> plt.show()
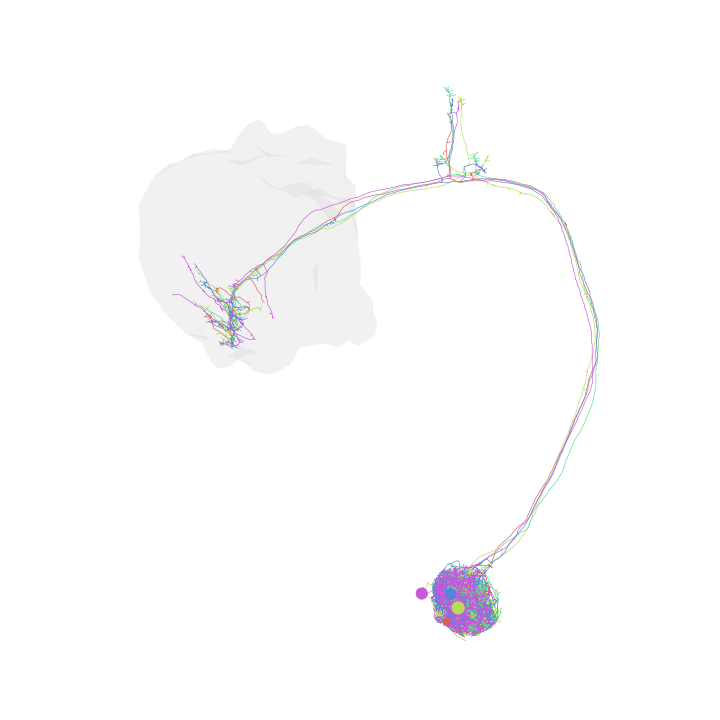
Change neuron colors
>>> fig, ax = navis.plot2d(nl, ... method='2d', ... view=('x', '-y'), ... color=['r', 'g', 'b', 'm', 'c', 'y']) >>> plt.show()
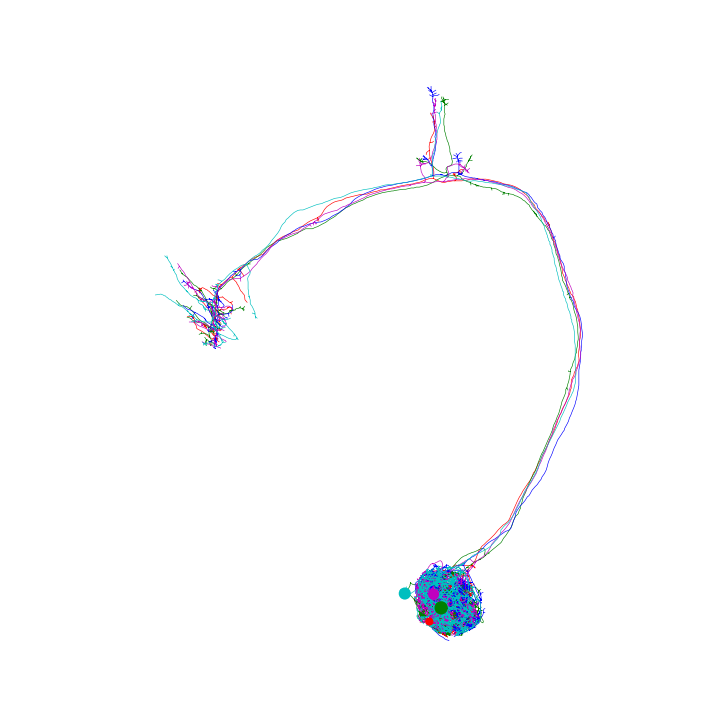
Plot in “fake” 3D
>>> fig, ax = navis.plot2d(nl, method='3d') >>> plt.show() >>> # In an interactive window you can dragging the plot to rotate
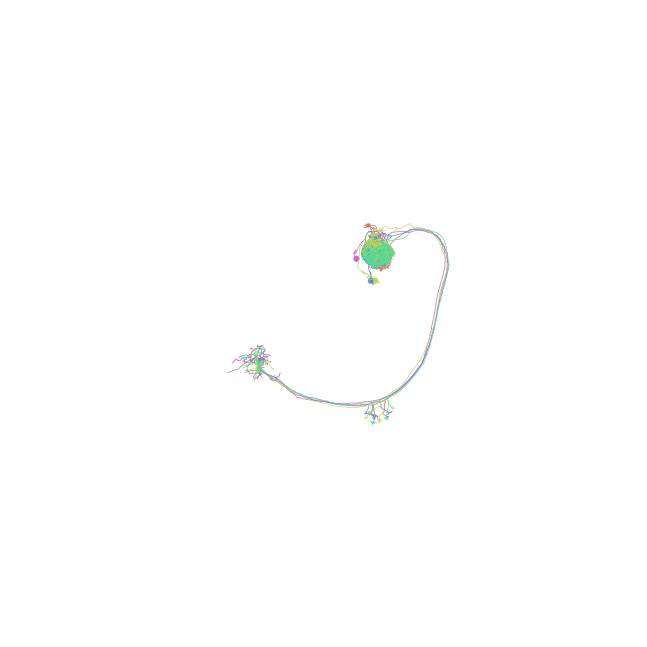
Plot in “fake” 3D and change perspective
>>> fig, ax = navis.plot2d(nl, method='3d') >>> # Change view to frontal (for example neurons) >>> ax.azim = ax.elev = 90 >>> # Change view to lateral >>> ax.azim, ax.elev = 180, 180 >>> ax.elev = 0 >>> # Change view to top >>> ax.azim, ax.elev = 90, 180 >>> # Tilted top view >>> ax.azim, ax.elev = -130, -150 >>> # Move camera >>> ax.dist = 6 >>> plt.show()
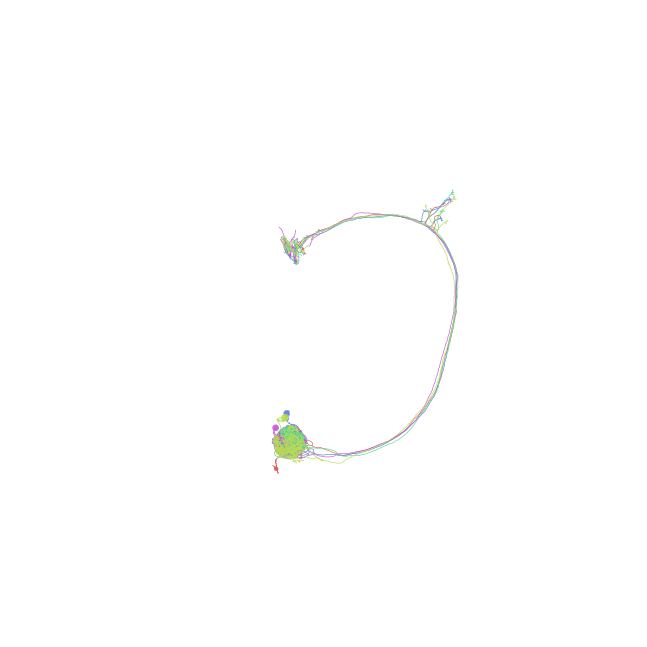
Plot using depth-coloring
>>> fig, ax = navis.plot2d(nl, method='3d', depth_coloring=True) >>> plt.show()
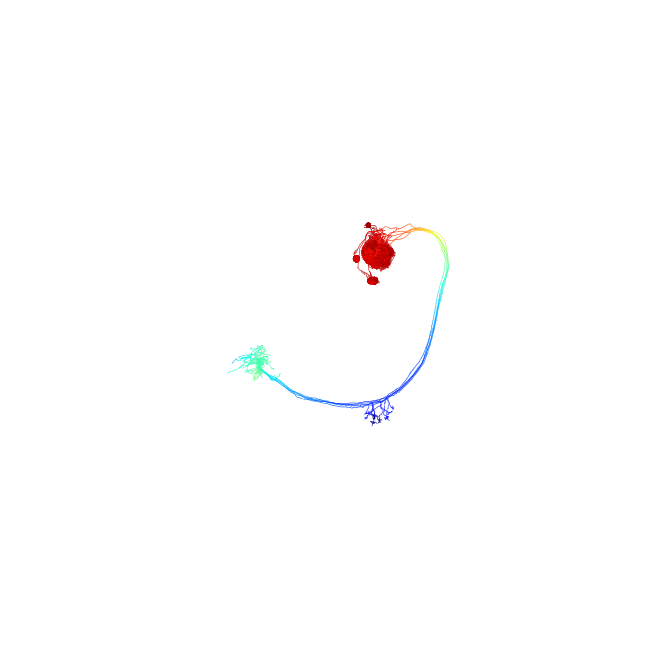
To close all figures
>>> plt.close('all')
See the plotting tutorial for more examples.
- Returns:
fig, ax
- Return type:
matplotlib figure and axis object
See also
navis.plot3d()Use this if you want interactive, perspectively correct renders and if you don’t need vector graphics as outputs.
navis.plot1d()A nifty way to visualise neurons in a single dimension.
navis.plot_flat()Plot neurons as flat structures (e.g. dendrograms).