Neuromorpho¶
Neuromorpho is a searchable online repository for neuron morphologies. At the time of writing it contains over 168k neurons from 91 different species. While the website features a comprehensive search and some neat stats, it can be useful to download these data to run your own analyses or compare to other data sets.
For that purpose, navis provides an interface to neuromorpho that wraps some of their API.
# Import navis
import navis
# Import the actual neuromorpho interface
import navis.interfaces.neuromorpho as nm
First we can use navis.interfaces.neuromorpho.get_neuron_fields() to figure out what we can search for:
fields = nm.get_neuron_fields()
fields[:10]
['neuron_id',
'neuron_name',
'archive',
'age_scale',
'gender',
'reference_pmid',
'reference_doi',
'note',
'age_classification',
'brain_region']
Next, we will use navis.interfaces.neuromorpho.get_available_field_values() to get an idea of the possible values for a given field:
species = nm.get_available_field_values('species')
species[:10]
['mouse',
'rat',
'drosophila melanogaster',
'human',
'zebrafish',
'monkey',
'chimpanzee',
'Xenopus laevis',
'Semipalmated sandpiper',
'C. elegans']
Let’s find a couple human neurons, shall we?
# Note that we're only fetching the first couple results
info = nm.find_neurons(species='human', page_limit=10)
info.head()
| neuron_id | neuron_name | archive | note | age_scale | gender | age_classification | brain_region | cell_type | species | ... | slicing_thickness | min_age | max_age | min_weight | max_weight | png_url | reference_pmid | reference_doi | physical_Integrity | _links | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 102399 | H17-03-010-11-13-01_656411100_m | Allen Cell Types | Year | Female | adult | [neocortex, occipital, middle temporal gyrus] | [interneuron, Aspiny] | human | ... | 350 | 38.0 | 38.0 | Not Reported | Not Reported | http://neuromorpho.org/images/imageFiles/Allen... | [27810003] | [10.1016/j.neuron.2016.10.019] | Dendrites Complete, Axon Incomplete | {'self': {'href': 'http://neuromorpho.org/api/... | |
| 1 | 102400 | H17-03-010-11-13-06_651089035_m | Allen Cell Types | Year | Female | adult | [neocortex, occipital, middle temporal gyrus] | [principal cell, Spiny] | human | ... | 350 | 38.0 | 38.0 | Not Reported | Not Reported | http://neuromorpho.org/images/imageFiles/Allen... | [27810003] | [10.1016/j.neuron.2016.10.019] | Dendrites Complete, Axon Incomplete | {'self': {'href': 'http://neuromorpho.org/api/... | |
| 2 | 102401 | H17-03-011-11-04-05_650978964_m | Allen Cell Types | Year | Male | adult | [neocortex, occipital, middle temporal gyrus] | [principal cell, Spiny] | human | ... | 350 | 30.0 | 30.0 | Not Reported | Not Reported | http://neuromorpho.org/images/imageFiles/Allen... | [27810003] | [10.1016/j.neuron.2016.10.019] | Dendrites & Axon Moderate | {'self': {'href': 'http://neuromorpho.org/api/... | |
| 3 | 102402 | H17-03-011-11-09-04_648981937_m | Allen Cell Types | Year | Male | adult | [neocortex, occipital, middle temporal gyrus] | [principal cell, Spiny] | human | ... | 350 | 30.0 | 30.0 | Not Reported | Not Reported | http://neuromorpho.org/images/imageFiles/Allen... | [27810003] | [10.1016/j.neuron.2016.10.019] | Dendrites & Axon Moderate | {'self': {'href': 'http://neuromorpho.org/api/... | |
| 4 | 102403 | H17-06-005-12-15-01_605485782_m | Allen Cell Types | Year | Male | adult | [neocortex, occipital, middle temporal gyrus] | [interneuron, Aspiny] | human | ... | 350 | 38.0 | 38.0 | Not Reported | Not Reported | http://neuromorpho.org/images/imageFiles/Allen... | [27810003] | [10.1016/j.neuron.2016.10.019] | Dendrites & Axon Complete | {'self': {'href': 'http://neuromorpho.org/api/... |
5 rows × 45 columns
Now on to fetching some actual neurons! The way you typically want to do this is by passing the info for your neurons of interest to navis.interfaces.neuromorpho.get_neuron():
neurons = nm.get_neuron(info)
neurons
102701 generated an exception: 404 Client Error: Not Found for url: http://neuromorpho.org/dableFiles/ortega/CNG%20version/WT6_02.CNG.swc
102706 generated an exception: 404 Client Error: Not Found for url: http://neuromorpho.org/dableFiles/ortega/CNG%20version/WT4_02.CNG.swc
102709 generated an exception: 404 Client Error: Not Found for url: http://neuromorpho.org/dableFiles/ortega/CNG%20version/WT9_02.CNG.swc
102719 generated an exception: 404 Client Error: Not Found for url: http://neuromorpho.org/dableFiles/ortega/CNG%20version/WT7_02.CNG.swc
102726 generated an exception: 404 Client Error: Not Found for url: http://neuromorpho.org/dableFiles/ortega/CNG%20version/WT5_02.CNG.swc
102728 generated an exception: 404 Client Error: Not Found for url: http://neuromorpho.org/dableFiles/ortega/CNG%20version/WT8_02.CNG.swc
| type | name | id | n_nodes | n_connectors | n_branches | n_leafs | cable_length | soma | units | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | navis.TreeNeuron | H17-03-010-11-13-01_656411100_m | 102399 | 1498 | None | 15 | 20 | 1684.614746 | [1, 2, 3] | 1 dimensionless |
| 1 | navis.TreeNeuron | H17-03-010-11-13-06_651089035_m | 102400 | 10451 | None | 69 | 81 | 13138.289062 | [1, 2, 3] | 1 dimensionless |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 492 | navis.TreeNeuron | H9-MECP2-T158M-T158M-CREB-17 | 103393 | 137 | None | 14 | 20 | 326.003082 | [1, 2, 3] | 1 dimensionless |
| 493 | navis.TreeNeuron | H9-MECP2-T158M-T158M-CREB-38 | 103394 | 172 | None | 43 | 52 | 324.924866 | [1, 2, 3] | 1 dimensionless |
As you can see some of these neurons are missing an SWC. That happens occasionally.
Let’s plot some of these!
import matplotlib.pyplot as plt
import seaborn as sns
fig, axes = plt.subplots(1, 10, figsize=(15, 5))
for n, c, ax in zip(neurons, sns.color_palette('muted', len(axes)), axes):
_ = navis.plot2d(n, method='2d', ax=ax, c=c)
ax.set_aspect('equal')
ax.set_axis_off()
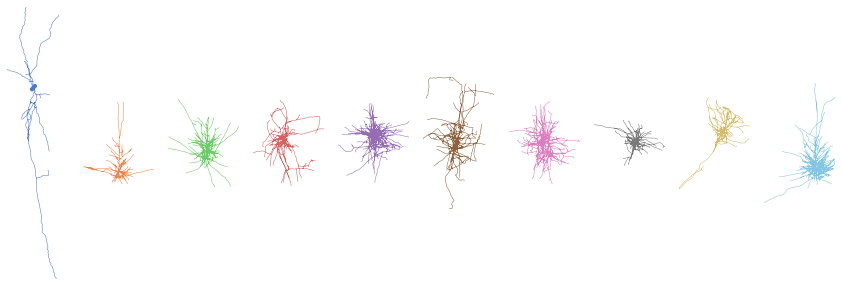
That’s all for now!